Targeted DamID (TaDa)
Targeted DamID (TaDa) (Southall, et al., 2013) enables cell-type specific profiling of DNA-binding proteins with fine spatial and temporal resolution. TaDa is based on DamID (van Steensel and Henikoff, 2000), a method for profiling protein binding which takes advantage of the lack of significant adenine methylation in eukaryotes. By fusing a bacterial DNA adenine methylase (Dam) to any protein of interest, only the regions bound by the protein will be methylated at the sequence GATC. Methylated GATC fragments are then selectively amplified via methylation-sensitive digestion and ligation-mediated PCR. By sequencing these fragments, the binding profile of the protein of interest can be obtained. A comparison of the method to ChIP-seq is shown below:
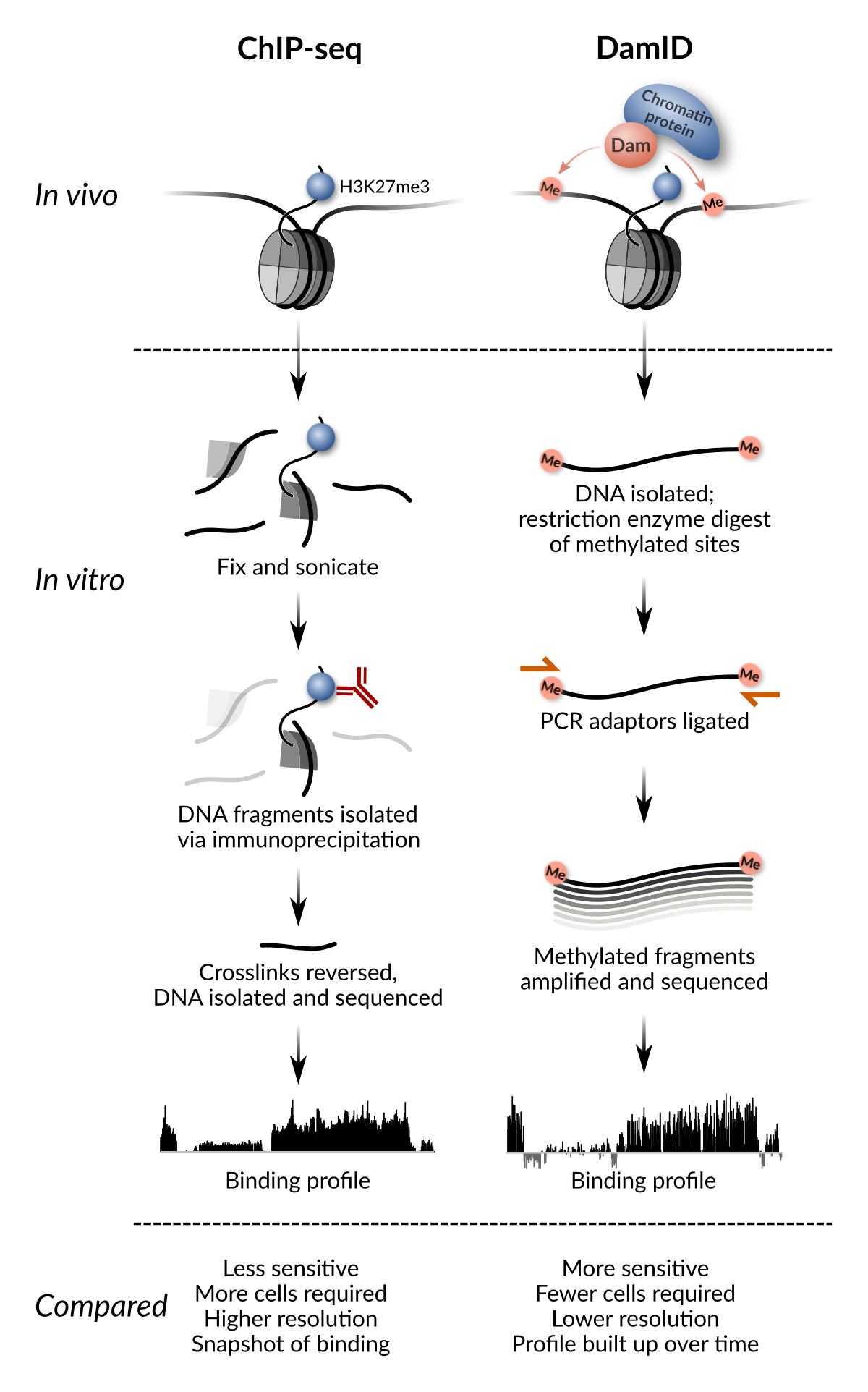
ChIP-seq and DamID compared (adapted from Fig. 1 of Delandre & Marshall 2019)
TaDa combines DamID with the GAL4 system in Drosophila, allowing Dam-fusion proteins to be expressed only in specific cell-types at precise temporal windows. TaDa is simple, robust, sensitive and highly reproducible, and works over a wide variety of different cell types and conditions. A further advantage of TaDa is that Dam-fusion proteins are expressed at almost undetectable levels such that cell fate remains unaffected.
TaDa protocol
Our current lab protocol for TaDa can be found in our recent chapter in Methods in Molecular Biology. Our current in-house protocol (version 2021-10-24) lists a few additional tips and tricks for NGS homebrew.
These protocols are an evolution from our earlier published work in Southall et al., Dev cell, 2013; Marshall and Brand, Bioinformatics, 2015; Marshall et al., Nature Protocols, 2016. If you’re just starting your TaDa journey, those papers may also be useful.
Something that we don’t tend to discuss in our published methods: TaDa is also extraordinarily easy to use. It requires no antibody purification steps or cell sorting, is highly reproducible and works pretty much every single time. TaDa requires only the most basic familiarity in molecular biology to perform: if you’ve done a restriction enzyme digest and a PCR reaction before, you already know all you need to perform TaDa.
TaDa vectors
Our new TaDa vectors pTaDaG, pTaDaG2 and pTaDaM use membrane-bound GFP or mCherry as the primary ORF, and as such clearly label the driving pattern within any TaDa experiment.
pTaDaG2 forms the backbone of pFlyORF-TaDa for our FlyORF-TaDa system.
pTaDaM forms the backbone for a membrane-bound fluorophore version of NanoDam. Our version of NanoDam also uses the transformer NLS for improved signal.
Vectors and Dam-only control flies are available upon request; if you find them useful, please cite the preprint above.
TaDa extensions
Originally, TaDa required cloning and transgenesis for every new protein to be profiled. However, recent extensions of the technique allow rapid profiling using existing fly resources.
FlyORF-TaDa allows existing FlyORF lines to be converted to TaDa lines with a simple cross.
NanoDam couples Dam to an anti-GFP nanobody to allow profiling of any GFP-fusion line with a simple cross.
Notes on data representation
DamID datasets are somewhat different to ChIP datasets. In general, we recommend that data is presented:
- As a log2(Dam-fusion/Dam) ratio
- At GATC fragment resolution
- Without obscuring or removing negative peaks
If datasets are compared between different cell types (and/or different conditions), scaling of the data (by dividing each dataset by its standard deviation) is appropriate before comparison. In general, it’s worth noting that DamID data is at most semi-quantitative. Direct quantitative comparisons are best made when using identical drivers and conditions.