FlyORF-TaDa
FlyORF-TaDa (Aughey et al, G3, 2021) allows the rapid generation of fly lines for profiling transcription factor binding with TaDa. The method was developed in collaboration with the Southall group at Imperial College, UK.
FlyORF-TaDa lines
The FlyORF-TaDa lines are now available for order from Bloomington.
| Stock No. | Genotype | Usage |
|---|---|---|
| 91637 | w[1118]; M{RFP[3xP3.PB] w[+mC]=UAS-flyORF.TaDa}ZH-86Fb | The base line to use as a Dam-only control (required for all experiments). |
| 91638 | w[1118]; P{y[+t7.7] w[+mC]=hs-FLPD5}attP40; M{RFP[3xP3.PB] w[+mC]=UAS-flyORF.TaDa}ZH-86Fb | The cassette-exchange-ready version of the line with hs-FlpD5 -- use this to make your new TaDa lines. |
Compatible FlyORF lines
FlyORF-TaDa works with all FlyORF lines generated in the ZH-86FB landing site and which have a compatible FRT5 site. Those are most of the available lines, but be aware when ordering there are some incompatible ones. (Incompatible lines to be avoided are a few early lines created with a plasmid that lacked the FRT5 site, as well as a growing library of “CC” lines for BiFC experiments inserted in a different site on Chr II.)
We also note that in our experience a few lines that should be compatible never throw recombinants. We are unsure why at this stage – they may be mislabelled in the database, or there may be occasional mutations in the FRT site.
The following types of lines in the FlyORF database should work with FlyORF-TaDa. We recommend using the ORF-3xHA lines in preference if available:
| FlyORF line type | FlyORF plasmid | Notes |
|---|---|---|
| ORF-3xHA | pGW-HA.attB | The majority of FlyORF lines are generated with this plasmid. Use these if available. |
| ORF-VN | pGW-HA.attB + in vivo tag swap | ORF lines with the N-terminal (1-154aa) of mVenus added at the C-terminus of the ORF. These should in theory work, but the C-terminal tag may cause aberrant binding. See Bischof, et al, Elife, 2018 for details. |
| ORF-VNshort | pGW-HA.attB + in vivo tag swap | As per the VN lines above, but the VNshort versions have a shorter linker between the ORF and the tag than the “VN” lines. See Bischof, et al, Elife, 2018 for details. |
Technical details
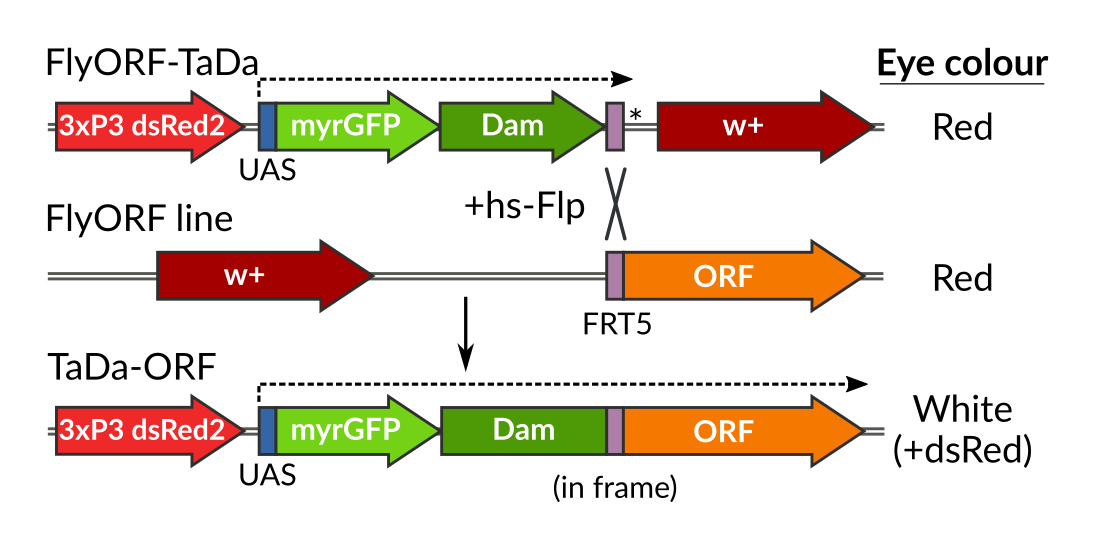
FlyORF-TaDa takes the existing FlyORF library of fly lines and uses a Flippase-mediated cassette exchange to convert them to Targeted DamID (TaDa) lines:
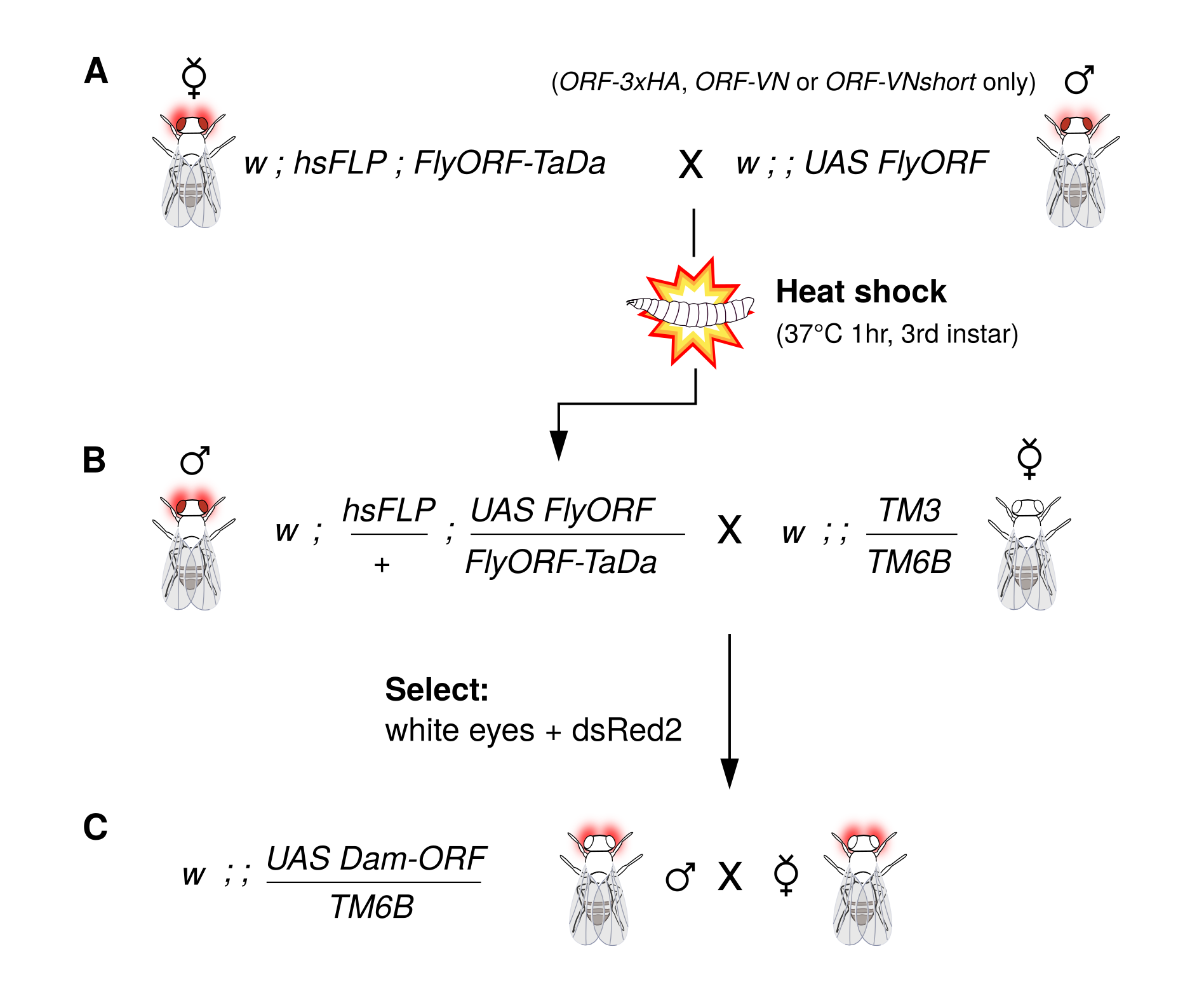
There are FlyORF lines for three quarters of all TFs and chromatin-modifying proteins in the Drosophila genome, so if you’ve got a favourite TF out there and want to profile where it binds cell-type specifically, the results are now just a cross away. We recommend the following crossing scheme to generate new lines:
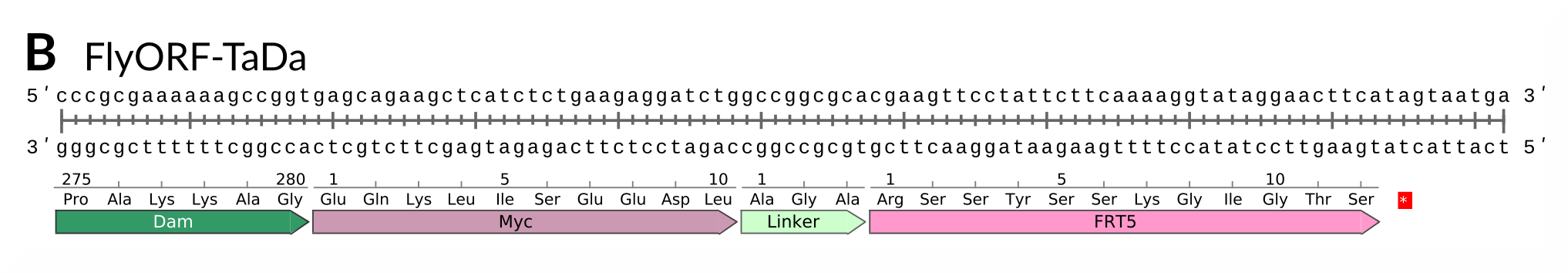
How does it work? The FlyORF library was neatly designed with an FRT5 site for flippase-mediated cassette-exchange. Our donor FlyORF-TaDa line has the TaDa system upstream of an FRT5 site, followed by a stop codon. By itself, the line acts as a Dam-only control:
And after Flippase-mediated conversion, the FRT5 site becomes a handy linker peptide:
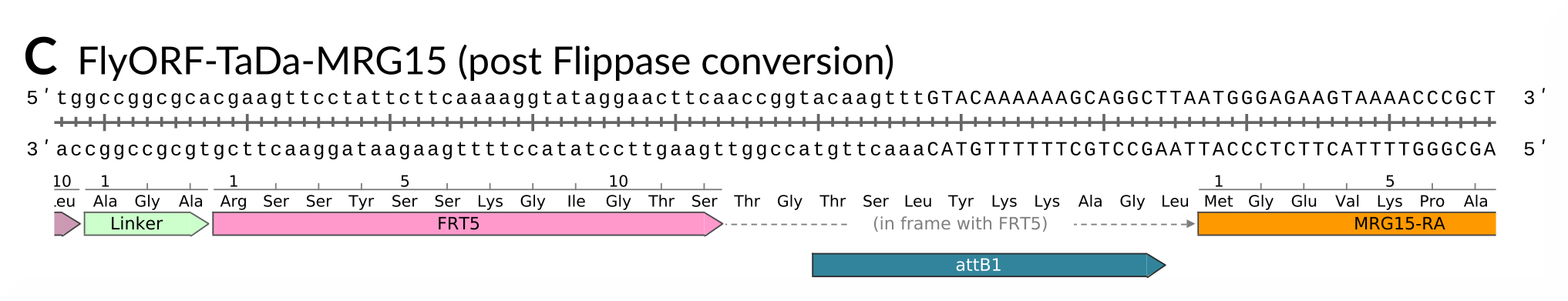
As an added bonus, you get an updated TaDa system with myrGFP as the primary open-reading frame, as per our TaDaG2 lines, so every experiment contains its own membrane-bound reporter. There’s now no excuse for not checking your drivers : )