Load genome-wide binding data and associated peak files or GRanges objects
Source:R/load_data.R
load_data_peaks.RdReads DamID-seq log2 ratio binding data either from bedGraph files or directly from a list of GRanges objects, and associated peak regions either from GFF/bed files or from a list of GRanges objects. This function is suitable for transcription factor binding analyses. For peak discovery, use an external peak caller (e.g. 'find_peaks').
Usage
load_data_peaks(
binding_profiles_path = NULL,
peaks_path = NULL,
binding_profiles = NULL,
peaks = NULL,
drop_samples = NULL,
maxgap_loci = 1000,
quantile_norm = FALSE,
organism = "drosophila melanogaster",
ensdb_genes = NULL,
BPPARAM = BiocParallel::bpparam(),
plot_diagnostics = interactive()
)Arguments
- binding_profiles_path
Character vector. Path(s) to directories or file globs containing log2 ratio binding tracks in bedGraph format. Wildcards ('*') supported.
- peaks_path
Character vector. Path(s) to directories or file globs containing the peak calls in GFF or BED format.
- binding_profiles
List of GRanges objects with binding profiles, one per sample.
- peaks
List of GRanges objects representing peak regions.
- drop_samples
A character vector of sample names or patterns to remove. Matching samples are removed from the analysis before normalisation and occupancy calculation. This can be useful for excluding samples that fail initial quality checks. Default: `NULL` (no samples are dropped).
- maxgap_loci
Integer, the maximum bp distance between a peak boundary and a gene to associate that peak with the gene. Default: 1000.
- quantile_norm
Logical (default: FALSE). If TRUE, quantile-normalise the signal columns across all datasets.
- organism
Organism string (lower case) to obtain genome annotation from (if not providing a custom `ensdb_genes` object) Default: "drosophila melanogaster".
- ensdb_genes
GRanges object: gene annotation. Automatically obtained from `organism` if NULL.
- BPPARAM
BiocParallel function (defaults to BiocParallel::bpparam())
- plot_diagnostics
Logical. If `TRUE` (the default in interactive sessions), diagnostic plots (PCA and correlation heatmap) will be generated and displayed for both the raw binding data and the summarised occupancy data.
Value
A list with components:
- binding_profiles_data
data.frame: Signal matrix for all regions, with columns chr, start, end, sample columns.
- peaks
list(GRanges): All loaded peak regions from input files or directly supplied.
- pr
GRanges: Reduced (union) peak regions across samples.
- occupancy
data.frame: Binding values summarised over reduced peaks, with overlap annotations.
- test_category
Character scalar; will be "bound".
Details
One of `binding_profiles_path` or `binding_profiles` must be provided. Similarly, one of `peaks_path` or `peaks` must be provided.
When supplying GRanges lists, each GRanges should contain exactly one numeric metadata column representing the binding signal, and all GRanges should be supplied as a named list, with element names used as sample names.
Examples
# Create a mock GRanges object for gene annotation
# This object, based on the package's unit tests, avoids network access
# and includes a very long gene to ensure overlaps with sample data.
mock_genes_gr <- GenomicRanges::GRanges(
seqnames = S4Vectors::Rle("2L", 7),
ranges = IRanges::IRanges(
start = c(1000, 2000, 3000, 5000, 6000, 7000, 8000),
end = c(1500, 2500, 3500, 5500, 6500, 7500, 20000000)
),
strand = S4Vectors::Rle(GenomicRanges::strand(c("+", "-", "+", "+", "-", "-", "+"))),
gene_id = c("FBgn001", "FBgn002", "FBgn003", "FBgn004", "FBgn005", "FBgn006", "FBgn007"),
gene_name = c("geneA", "geneB", "geneC", "geneD", "geneE", "geneF", "LargeTestGene")
)
# Get path to sample data files included with the package
data_dir <- system.file("extdata", package = "damidBind")
# Run loading function using sample files and mock gene annotations
loaded_data <- load_data_peaks(
binding_profiles_path = data_dir,
peaks_path = data_dir,
ensdb_genes = mock_genes_gr,
quantile_norm = TRUE
)
#> Locating binding profile files
#> Building binding profile dataframe from input files ...
#> - Loaded: Bsh_Dam_L4_r1-ext300-vs-Dam.kde-norm
#> - Loaded: Bsh_Dam_L4_r2-ext300-vs-Dam.kde-norm
#> - Loaded: Bsh_Dam_L5_r1-ext300-vs-Dam.kde-norm
#> - Loaded: Bsh_Dam_L5_r2-ext300-vs-Dam.kde-norm
#> Locating peak files
#> Applying quantile normalisation
#> Calculating occupancy over peaks
#> Calculating average occupancy for 1208 regions...
#> Generating diagnostic plots...
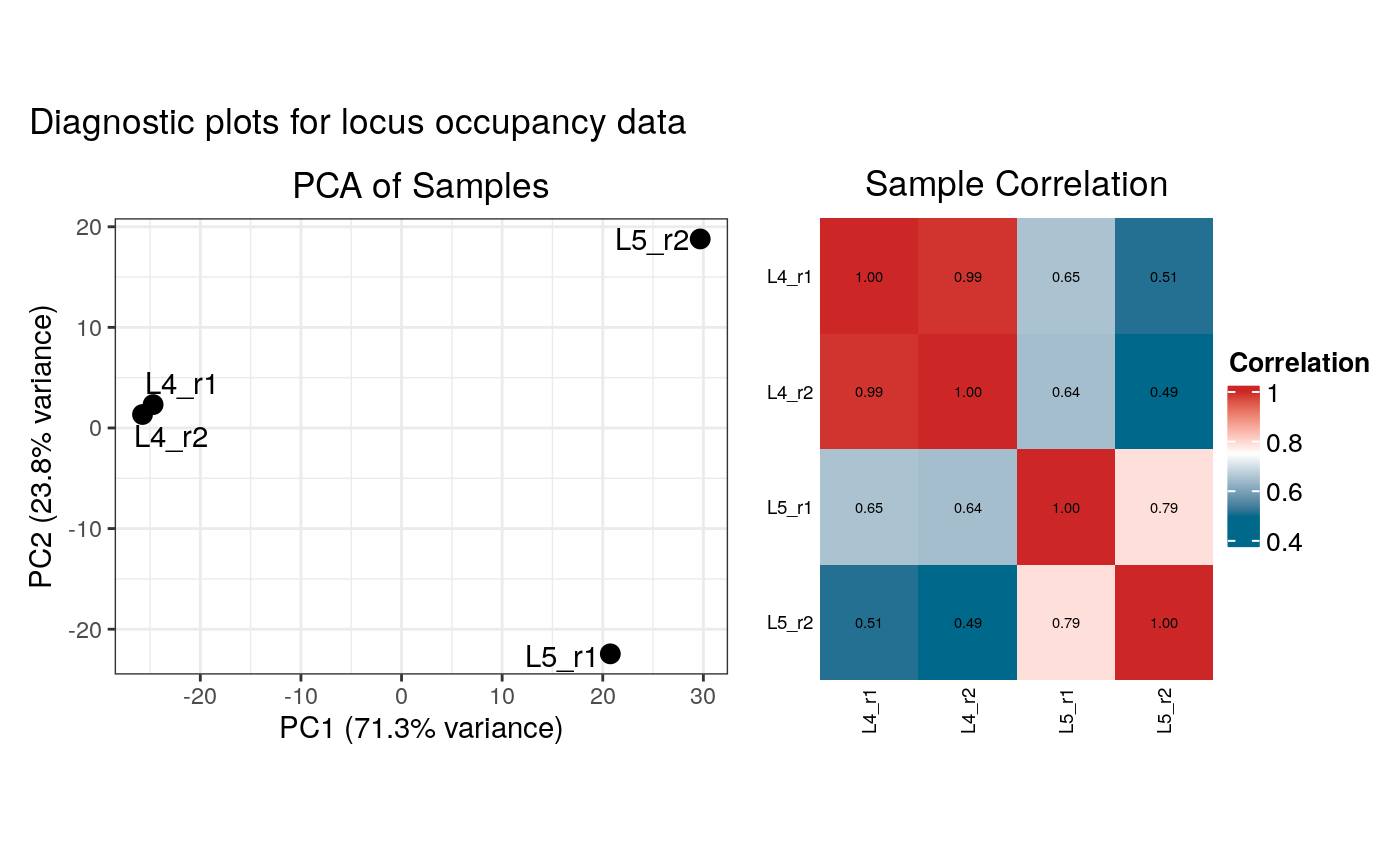
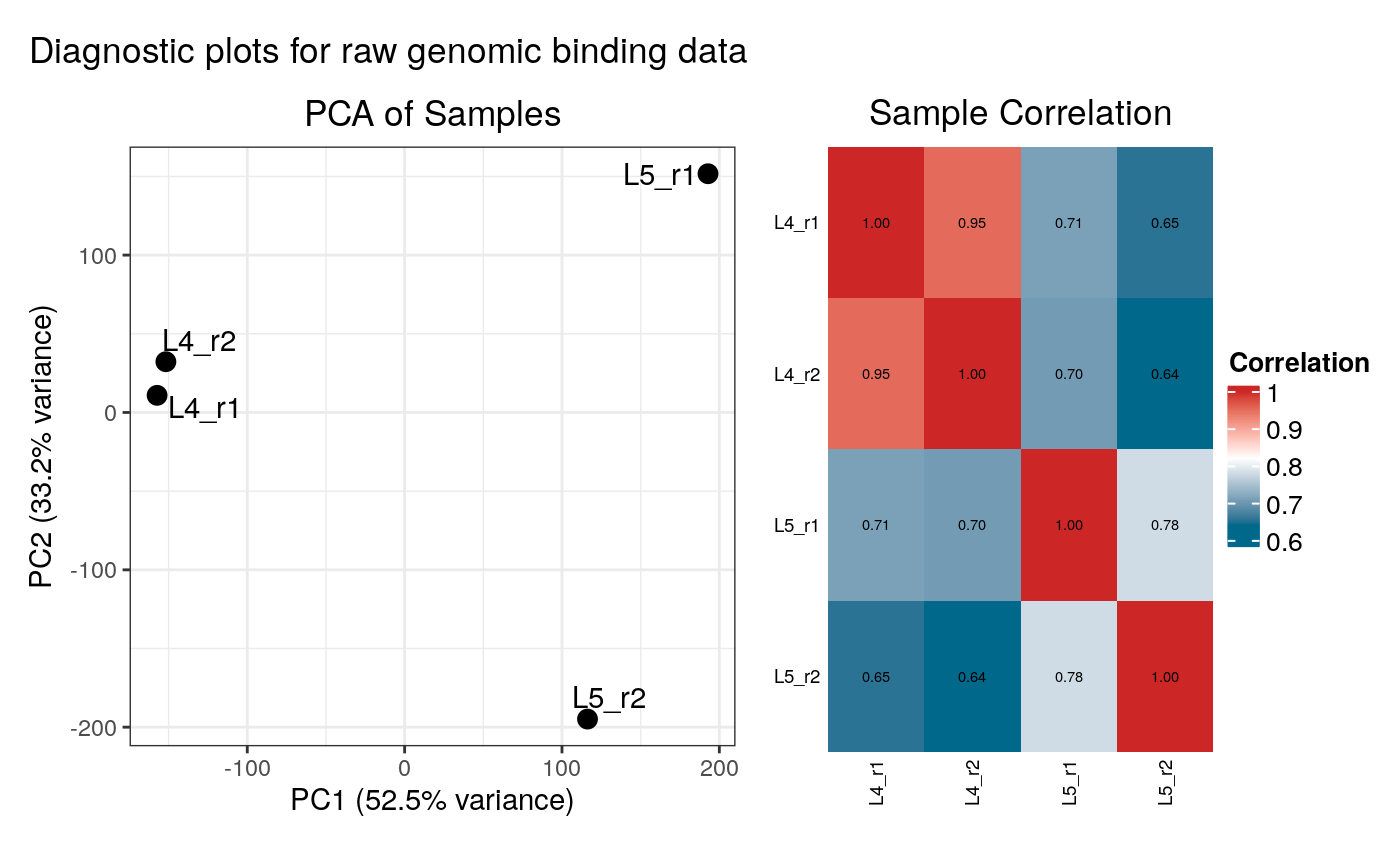 # View the structure of the output
str(loaded_data, max.level = 1)
#> List of 5
#> $ binding_profiles_data:Formal class 'GRanges' [package "GenomicRanges"] with 7 slots
#> $ peaks :List of 4
#> $ pr :Formal class 'GRanges' [package "GenomicRanges"] with 7 slots
#> $ occupancy :'data.frame': 1208 obs. of 9 variables:
#> $ test_category : chr "bound"
# View the structure of the output
str(loaded_data, max.level = 1)
#> List of 5
#> $ binding_profiles_data:Formal class 'GRanges' [package "GenomicRanges"] with 7 slots
#> $ peaks :List of 4
#> $ pr :Formal class 'GRanges' [package "GenomicRanges"] with 7 slots
#> $ occupancy :'data.frame': 1208 obs. of 9 variables:
#> $ test_category : chr "bound"